Modelica Performance Analyzer¶
A common problem when simulating models in an equation-based language like Modelica is that the model may contain non-linear equation systems. These are solved in each time-step by extrapolating an initial guess and running a non-linear system solver. If the simulation takes too long to simulate, it is useful to run the performance analysis tool. The tool has around 5~25% overhead, which is very low compared to instruction-level profilers (30x-100x overhead). Due to being based on a single simulation run, the report may contain spikes in the charts.
When running a simulation for performance analysis, execution times of user-defined functions as well as linear, non-linear and mixed equation systems are recorded.
To start a simulation in this mode, turn on profiling with the following command line flag >>> setCommandLineOptions("--profiling=all")
The generated report is in HTML format (with images in the SVG format), stored in a file modelname_prof.html, but the XML database and measured times that generated the report and graphs are also available if you want to customize the report for comparison with other tools.
Below we use the performance profiler on the simple model A:
model ProfilingTest
function f
input Real r;
output Real o = sin(r);
end f;
String s = "abc";
Real x = f(x) "This is x";
Real y(start=1);
Real z1 = cos(z2);
Real z2 = sin(z1);
equation
der(y) = time;
end ProfilingTest;
We simulate as usual, after setting the profiling flag:
>>> setCommandLineOptions("--profiling=blocks+html")
true
>>> simulate(ProfilingTest)
record SimulationResult
resultFile = "«DOCHOME»/ProfilingTest_res.mat",
simulationOptions = "startTime = 0.0, stopTime = 1.0, numberOfIntervals = 500, tolerance = 1e-6, method = 'dassl', fileNamePrefix = 'ProfilingTest', options = '', outputFormat = 'mat', variableFilter = '.*', cflags = '', simflags = ''",
messages = "LOG_SUCCESS | info | The initialization finished successfully without homotopy method.
LOG_SUCCESS | info | The simulation finished successfully.
Warning: empty y range [3:3], adjusting to [2.97:3.03]
Warning: empty y range [3:3], adjusting to [2.97:3.03]
Warning: empty y range [2:2], adjusting to [1.98:2.02]
Warning: empty y range [2:2], adjusting to [1.98:2.02]
Warning: empty y range [3:3], adjusting to [2.97:3.03]
Warning: empty y range [3:3], adjusting to [2.97:3.03]
LOG_STDOUT | info | Time measurements are stored in ProfilingTest_prof.html (human-readable) and ProfilingTest_prof.xml (for XSL transforms or more details)
",
timeFrontend = 0.001839541,
timeBackend = 0.015235895000000001,
timeSimCode = 0.00156207,
timeTemplates = 0.004443672,
timeCompile = 0.622192725,
timeSimulation = 0.054278719,
timeTotal = 0.699714782
end SimulationResult;
"Warning: There are nonlinear iteration variables with default zero start attribute found in NLSJac0. For more information set -d=initialization. In OMEdit Tools->Options->Simulation->Show additional information from the initialization process, in OMNotebook call setCommandLineOptions(\"-d=initialization\").
Warning: The initial conditions are not fully specified. For more information set -d=initialization. In OMEdit Tools->Options->Simulation->Show additional information from the initialization process, in OMNotebook call setCommandLineOptions(\"-d=initialization\").
"
Profiling information for ProfilingTest¶
Information¶
All times are measured using a real-time wall clock. This means context switching produces bad worst-case execution times (max times) for blocks. If you want better results, use a CPU-time clock or run the command using real-time priviliges (avoiding context switches).
Note that for blocks where the individual execution time is close to the accuracy of the real-time clock, the maximum measured time may deviate a lot from the average.
For more details, see ProfilingTest_prof.xml.
Settings¶
Name |
Value |
|---|---|
Integration method |
dassl |
Output format |
mat |
Output name |
|
Output size |
24.0 kB |
Profiling data |
|
Profiling size |
0 B |
Summary¶
Task |
Time |
Fraction |
|---|---|---|
Pre-Initialization |
0.000166 |
5.90% |
Initialization |
0.000613 |
21.79% |
Event-handling |
0.000000 |
0.00% |
Creating output file |
0.000164 |
5.83% |
Linearization |
NaN% |
|
Time steps |
0.001530 |
54.39% |
Overhead |
0.000137 |
4.87% |
Unknown |
NaN |
NaN% |
Total simulation time |
0.002813 |
100.00% |
Global Steps¶
Steps |
Total Time |
F raction |
Average Time |
Max Time |
De viation |
|
|---|---|---|---|---|---|---|
499 |
0 .001530 |
54.39% |
3.0661 3226452 906e-06 |
0.00 0069961 |
21.82x |
Measured Function Calls¶
Name |
Calls |
Time |
F raction |
Max Time |
De viation |
|
|---|---|---|---|---|---|---|
|Graph th umbnail f unction fun0|
th umbnail count f unction fun0| |
` Profili ngTest. f <#lin e=0>`__ |
1512 |
0.00 0031019 |
1.10% |
0.00 0000361 |
16.60x |
Measured Blocks¶
Name |
Calls |
Time |
F raction |
Max Time |
De viation |
|
|---|---|---|---|---|---|---|
th umbnail count eq0| |
` <# eq0>`__ |
8 |
0.00 0097126 |
3.45% |
0.00 0097261 |
7.01x |
th umbnail count eq11| |
` <#e q11>`__ |
4 |
0.00 0001803 |
0.06% |
0.00 0001861 |
3.13x |
th umbnail count eq19| |
` <#e q19>`__ |
1006 |
0.00 0535814 |
19.05% |
0.00 0019001 |
34.67x |
th umbnail count eq21| |
` <#e q21>`__ |
1508 |
0.00 0575477 |
20.46% |
0.00 0027081 |
69.96x |
Equations¶
Name |
Variables |
|---|---|
eq0 |
|
eq1 |
|
eq2 |
|
eq3 |
|
eq4 |
|
eq5 |
` <#var0>`__ |
eq6 |
` <#var0>`__ |
eq7 |
` <#var0>`__ |
eq8 |
` <#var0>`__ |
eq9 |
|
eq10 |
|
eq11 |
|
eq12 |
|
eq13 |
|
eq14 |
|
eq15 |
` <#var0>`__ |
eq16 |
` <#var0>`__ |
eq17 |
` <#var0>`__ |
eq18 |
` <#var0>`__ |
eq19 |
|
eq20 |
|
eq21 |
|
eq22 |
|
eq23 |
Variables¶
Name |
Comment |
|---|---|
This is x |
|
This report was generated by OpenModelica on 2024-11-14 22:13:39.
Genenerated JSON for the Example¶
{
"name":"ProfilingTest",
"prefix":"ProfilingTest",
"date":"2024-11-14 22:13:39",
"method":"dassl",
"outputFormat":"mat",
"outputFilename":"ProfilingTest_res.mat",
"outputFilesize":24581,
"overheadTime":0.00013742,
"preinitTime":0.000165871,
"initTime":0.000612741,
"eventTime":0,
"outputTime":0.000163714,
"jacobianTime":5.84296e-06,
"totalTime":0.00281345,
"totalStepsTime":2.71072e-06,
"totalTimeProfileBlocks":0.00121022,
"numStep":499,
"maxTime":6.996072e-05,
"functions":[
{"name":"ProfilingTest.f","ncall":1512,"time":0.000031019,"maxTime":0.000000361}
],
"profileBlocks":[
{"id":0,"ncall":8,"time":0.000097126,"maxTime":0.000097261},
{"id":11,"ncall":4,"time":0.000001803,"maxTime":0.000001861},
{"id":19,"ncall":1006,"time":0.000535814,"maxTime":0.000019001},
{"id":21,"ncall":1508,"time":0.000575477,"maxTime":0.000027081}
]
}
Using the Profiler from OMEdit¶
When running a simulation from OMEdit, it is possible to enable profiling information, which can be combined with the transformations browser.
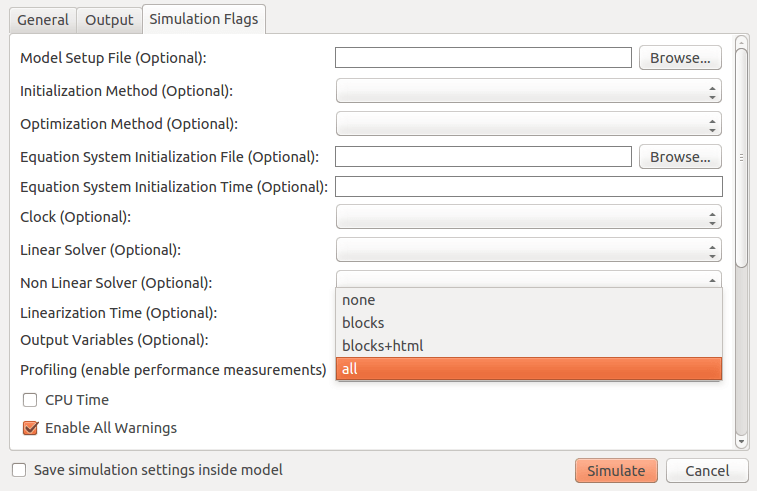
Figure 137 Setting up the profiler from OMEdit.¶
When profiling the DoublePendulum example from MSL, the following output in Figure 138 is a typical result. This information clearly shows which system takes longest to simulate (a linear system, where most of the time overhead probably comes from initializing LAPACK over and over).
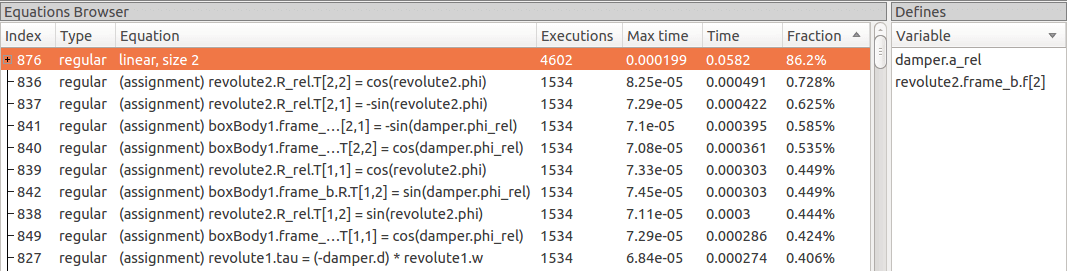
Figure 138 Profiling results of the Modelica standard library DoublePendulum example, sorted by execution time.¶
